|
Replication
DNA must be duplicated in a process called replication before a cell divides. The replication of
DNA allows each daughter cell to contain a full complement of chromosomes. One
of the single strands of the double-stranded DNA molecules is the template for the generation of the other.  DNA replication is semiconservative. Each
of the two molecules double-stranded molecules resulting from replication contains one old and one new single strand. DNA replication is semiconservative. Each
of the two molecules double-stranded molecules resulting from replication contains one old and one new single strand.
Proving How the DNA is Replicated
- Watson and Crick proposed the hypothesis of semiconservative replication
in their original Nature paper.
- What was left was to prove it.
- The Meselson Stahl experiment was a real classic in genetic research.
- It was a critical experiment by design. This means that no matter
what the result, they would have their answer.
Meselson and Stahl Experiment
- E. coli are grown in heavy nitrogen (15N) for
many generations.
- This caused the nitrogen in the DNA molecule of each cell to contain
15N, a heavier than typical isotope.
- The E. coli were then grown for one or two cell divisions
in 14N, the lighter and typical isotope.

-
- DNA was spun in a cesium chloride gradient. Meselson and Stahl
actually invented this technique, called density centrifugation, which now has many other applications, just for the purposes
of this experiment.
- The cesium chloride gradient and centrifugation separates molecules
based on their density.
- The DNA molecules with 15N are more dense than those
with 14N, and band below DNA with 14N.
- If two bands were observed after one division in 14N,
there would have been wholly old strands and wholly new strands. This would have been consistent with and meant the replication
was conservative.
- If there was just one band after one division, replication could
be either dispersive or semiconservative.
- The result was just one band after one division.
- If one or a long smear was observed after two divisions in 14N
containing medium, dispersive replication would have been the mode.
- If intermediate weight and light weight molecules were found,
semiconcervative would be the mode.
- This is what was found; the replication was semiconservative.
This was the predicted outcome of Watson and Crick.

DNA Replication: A Closer Look
- DNA polymerase enzymes are involved in repair and chromosome replication.
- Prokaryotic DNA I: Discovered by the famous Dr. Authur Kornberg
- this enzyme polymerizes small DNA segments during replication and repair. Most of the DNA polymerase found in prokaryotic
cells is DNA polymerase I.
- DNA III: This is the primary DNA polymerase involved in DNA replication
of bacterial chromosomes.
- DNA always replicates from 5' to 3' in direction using
the antiparallel strand as a template.
- Therefore, for each double-strand of DNA, one strand must replicate
in the opposite direction of the other.
- Why only in the 5' to 3' direction? Because a new base can only
be added to the 3' OH position.

Also, a new base can only be added to an existing polymer!
- So how does it start? (This
question is answered later in this unit.)
- A special DNA template dependent RNA polymerase called primase
makes a stretch of RNA that complements the template strand and forms a RNA / DNA duplex.
- Next, DNA polymerase adds to that RNA molecule.
- Later, the RNA molecule is removed.
- Initiation: this involves the assembly of a replication
fork (bubble) at an origin of replication sequence of DNA found at a specific site of the circular chromosome of a bacterium.
The fork is generated by a complex of proteins called a primosome.
- Elongation: this is the addition of bases by another
complex of proteins called the replisome. Parental strands unwind and daughter strands are synthesized.
- Termination: the duplicated chromosomes separate
from each other. This is actually a complicated procedure, the details of which we won't cover in this course.
How was DNA Replication Studied?
- Geneticists typically study biological processes by finding lack-of-function
mutants.
- Non-functional mutants for any necessary component of replication
die. Only by using conditional lethals could the process be dissected genetically.
- A temperature conditional mutant might stop replication at 42oC
for example.
- This is because unlike the normal version of the gene, these mutants
code for an enzyme that is inactive at a certain temperature.
- Cells are first grown in a permissive condition, and then are
moved into the inhibiting condition.
- Using this process, dna genes have been identified.
- Some are quick-stop mutants. These are involved in the elongation
process.
- Some are slow-stop mutants. These are involved in initiation or
termination.
- Some types of DNA polymerase are occasionally called DNA replicases.
- Other types are considered DNA repair enzymes.
- All DNA polymerases polymerize a polynucleotide by adding to an
existing double-stranded stretch of DNA.
- Aggregates of specific subunits of polypeptides makeup the replicase
molecule.
- In E. coli, there are three major DNA polymerases: DNA
polymerase I, II and III.
- DNA poly I is found in the highest concentration of all
DNA polymerases; it is involved in DNA repair and assists with primary DNA replication.
- DNA poly II is exclusively involved in repair.
- DNA poly III is the major DNA polymerase role.
- All DNA polymerases add to the 3’ OH of the existing polynuceotide.
- DNA polymerases have other enzymatic activities as well.
- One is an exonuclease activity, the digestion of the polymer
into monomers, which is the reverse activity of polymerization.
- One can diagram it to look a bit like PackMan of the video
game. If the enzyme has exonuclease activity, it works in the 3' to 5' direction, which is the opposite of the 5' to 3' polymerase
activity. Some DNA polymerase holoenzyme complexes have both a 3' to 5' and a 5' to 3' exonuclease activity. DNA polymerase
I does for example.
- This function is involved with error reduction:
"proof reading". It is also involved with removal of the RNA primers and replacement of the RNA with DNA. There is also a
lot of involvement of these activities in non-replicative DNA repair.

Another enzymatic activity associated with DNA polymerase is proofreading.
This is a check that the correct base was incorporated immediately after the fact.
- The accuracy or fidelity of DNA replication can be measured relatively
easily and has been for many different types of organisms.
- It can be expressed in a number of different ways, for example,
errors per nucleotide incorporated.
- It varies substantially between prokaryotes, eukaryotes, unicellular
and multicellular organisms.
- A typical value given by some genetics textbooks is 1 error/genome/1000
bacterial replications.
- It would be 10-3 instead of 10-8 to 10-10,
if there were no checks on the system.
- Checks include a pre-synthesis check to verify the incoming base
is correct.
- The proofreading check verifies that the correct base was incorporated.
- The 3’ to 5’ exonuclease activity assists when it
is found that the base incorporated was incorrect. It removes the incorrect base so that the polymerase can try again to get
it right.
- The DNA polymerase I enzyme is 103 kD.
- Of this a 68 kD portion is called the Klenow fragment.
- Klenow has the 3’ to 5’ exonuclease and the polymerase
activity in different regions of the fragment.
- Another 35 kD fragment has a 5’ to 3’ exonuclease
activity.
- The E. coli DNA polymerase I incorporates about 10 bases
in length at a time. This is called its processivity. Some organisms poly I incorporate more, some less.
- Nick translation is an extremely useful technique for labeling
DNA.
- First DNA is nicked using a shearing force such as forcing the
solution containing the DNA molecules through a fine bore hypodermic needle.
- The DNA with nicks, DNA polymerase I and labeled deoxiribonucleoside
triphosphates are combined in solution, and the DNA polymerase I molecules attach to the nick, use their 5’ to 3’
exonuclease activity to digest out existing nucleotides, while replacing them with labeled ones using their polymerase activity.
- This technique can be used to incorporate
radioactive labeled nucs.
- DNA polymerase III is the main polymerase for E. coli chromosome
replication.
- It is made up of many subunits.
- It has several different activities.
- It is found in different forms ( subunit associations).
- E. coli's has 3’ to 5’ exonuclease proofreading.
- The gene dna E codes for the 130 kD
fragment which has the polymerase activity.
- The alpha subunit works to increase error proofreading ability.

- Frameshift(s) are extra or missing base(s).
- Frameshifts are influenced by the processivity of the enzyme.
- Slippage is less if processivity is greater
(adding more bases before disassociating).

Strings of some bases, especially short repeats, can increase
the frequency of frameshift mistakes.
Substitutions are when the wrong nucleotide is incorporated.
Substitution rates are strongly influenced by the proofreading
function of the DNA polymerase.
Eukaryotic DNA Polymerase
- DNA Polymerase alpha and delta replicate the DNA.
- DNA polymerase alpha is associated with initiation, and delta
extends the nascent strands.
- DNA polymerase epsilon and beta are used for repair.
- DNA polymerase gamma is for replication of
mitochrondrial DNA.
- All DNA polymerases require a 3’ end of an existing double-stranded
molecule to add nucleotides to.
- There are several different ways this is done.
- Cells make RNA primers.
- Some viruses use a preformed RNA.
- A nick or gap in the phosphodiester backbone
of the DNA molecule is used as the starting point.
Semidiscontinuous synthesis
- The replication fork moves in one direction.
- However, DNA is replicated in the 5’ to 3’ direction
only.
- How can this be?
- There is a leading and lagging strand.
- Short pieces of DNA called Okazaki fragments
with a short stretch of RNA at each 5' end are found temporarily on the lagging strand.
- (The leading strand might at times appear to be fragmented
also, but this is due to deoxi-uracil getting mistakenly incorporated instead of thymine, and then the uracil getting cut
out by uracil glycosidase and AP endonuclease.)
- This system of replication is called semi-discontinuous because
one strand, the leading, is continuous, and the other, the lagging, is discontinuous.

- Each Okazaki fragment starts with about 10 bases of RNA primer.
- What happens to Okazaki fragments? Does chromosomal DNA really
contain RNA also?
- Ligase fails to seal gaps if the gap is between an RNA and a DNA
base, which is how the gap exists between each separate Okazaki fragment.
- Time and the gap give DNA polymerase the opportunity to do the
Pack Man routine.
- DNA polymerase I eats the RNA bases (exonuclease activity),
whilst incorporating new DNA bases (DNA polymerase activity).
- Actually, a DNA polymerase I molecule will digest and add a few
bases, and then fall off.
- Because a gap still exists, another DNA polymerase I molecule
will do the same again.
- If the gap is now between two deoxiribonucleotides, either DNA
polymerase I or ligase will associate with the gap.
- If ligase gets their first, it seals the gap.
- Ligase will "get their first" sooner or later, depending on concentrations,
and the gap will get sealed.
- DNA polymerase I is using the end of the last Okazaki fragment,
which is composed of DNA, as its primer.
- DNA polymerase I removes and adds bases beyond the RNA primer
stretch a bit, unnecessarily.
- Ligase seals the gap.
- Ligases use energy from NADH (E. coli)
or ATP (T4 phage).
- First, helicase separates the strands of the double-stranded DNA
molecule; E. coli's DnaB codes for helicase.
- Single-strand DNA binding proteins maintain the strands in this
region as single-stranded.
- This occurs at the oriC site, the bacterial origin of replication.
- Primase, the DNA template dependent, RNA polymerase, which makes
the primers, is coded for by the dna G gene.
- Unwinding the DNA by helicase requires ATP as an energy source
and substrate.
- A note on unwinding enzymes:
- Topoisomerases effect the coiling of double-stranded DNA.
- Type 1 work by making single-strand breaks; Type 2
work by making double-strand breaks.
- The replication fork moves in opposite directions away from the
origin of replication, forming two bubbles.
- How does it know when to stop in bacteria, so that just one round
of replication takes place, and you don't just get a jumble of many copies?
- In E. coli, there are ter elements, which are 23
base pair consensus sequences.
- These are binding sites for the tus gene protein.
- This stops the Dna B product, helicase, from unwinding
DNA in the ter element regions.

- Strands synthesize just up to ter.
- The strand extension is prevented in one direction
but can read through in the other direction until the bubbles meet. The two strands must then disconnect from each other.
-
Summary
- Only one strand can replicate continuously. The other is discontinuous.
- On the discontinuous strand, short segments are called Okazaki
Fragments, these are about 1,500 bases in length in prokaryotes, and 150 bases in eukaryotes.
- The continuous is called the leading strand, the discontinuous
is the lagging strand.
- The leading has high processivity, it continues until the entire
strand is replicated.
- The lagging requires these repeated steps:
- 1. Primer synthesis
- 2. Elongation
- 3. Primer removal

The average human chromosome contains 150 x 106 nucleotide pairs which are copied at about 50 base
pairs per second. The process would take a month (rather than the hour it actually does) but for the fact that there are many
places on the eukaryotic chromosome where replication can begin. Replication begins at some replication origins earlier in
S phase than at others, but the process is completed for all by the end of S phase. As replication nears completion, "bubbles"
of newly replicated DNA meet and fuse, finally forming two new molecules.
With their multiple origins, how does the eukaryotic cell know which origins have been already replicated
and which still await replication?
An observation: When a cell in G2 of the cell cycle is fused with a cell in S phase, the DNA of the G2 nucleus does not begin replicating again even
though replication is proceeding normally in the S-phase nucleus. Not until mitosis is completed, can freshly-synthesized
DNA be replicated again.
Two control mechanisms have been identified — one positive and one negative. This redundancy
probably reflects the crucial importance of precise replication to the integrity of the genome.
 In order to be replicated, each origin of replication must be bound by: In order to be replicated, each origin of replication must be bound by:
- an Origin Recognition Complex of proteins (ORC). These remain on the DNA throughout
the process.
- Accessory proteins called licensing factors. These accumulate in the nucleus during G1
of the cell cycle. They include:
- CDC-6 and CDT-1, which bind to the ORC and are essential for coating the DNA with
- MCM proteins. Only DNA coated with MCM proteins (there are 6 of them) can be replicated.
- Once replication begins in S phase,
- CDC-6 and CDT-1 leave the ORCs (the latter by ubiquination and destruction in proteasomes).
- The MCM proteins leave in front of the advancing replication fork.
G2 nuclei also contain at least one protein — called geminin — that prevents
assembly of MCM proteins on freshly-synthesized DNA (probably by sequestering Cdt1).
As the cell completes mitosis, geminin is degraded so the DNA of the two daughter cells will be able
to respond to licensing factors and be able to replicate their DNA at the next S phase.
Endoreplication is the replication of DNA during the S phase of the cell cycle without the subsequent completion of mitosis and/or cytokinesis. (Endoreplication is also known as endoreduplication.)
Endoreplication occurs in certain types of cells in both animals and plants. There are several variations:
- replication of DNA with completion of mitosis but no cytokinesis.
- repeated replication of DNA without forming new nuclei in telophase. This can result in:
- Polyploidy: the replicated chromosomes retain their individual identity.
- Polyteny: the replicated chromosomes remain in precise alignment forming "giant" chromosomes.
- various intermediate conditions between 1 and 2
Mitosis without cytokinesis produces a mass of cytoplasm with many nuclei. Some examples:
- the free-nuclei stage in the embryonic development of flies like Drosophila
- the plasmodial slime molds like Physarum polycephalum
Polyploidy
Cells are polyploid if they contain more than two haploid (n) sets of chromosomes; that is, their chromosome
number is some multiple of n greater than the 2n content of diploid cells. For example, triploid (3n)
and tetraploid cell (4n) cells are polyploid.
Polyploidy is usually limited to certain tissues in animals, such as
- liver cells;
- megakaryocytes; Megakaryocytes, from which platelets are made, may pass through as many as seven S phases producing a giant cell with a single nucleus containing
128n chromosomes. Its fragmentation yields platelets.
- giant trophoblast cells in the placenta.
Polyploidy is very common in plants.

The most thoroughly-studied examples of polyteny are the giant chromosomes found in certain cells of flies.
The photomicrograph (courtesy of B. P. Kaufmann) shows the polytene chromosomes in a salivary gland cell of
Drosophila melanogaster. Such chromosomes are found in other large, active cells as well.
- Each of Drosophila's 4 pairs of chromosomes has undergone 10 rounds of DNA replication.
- The maternal and paternal homologs — as well as all their duplicates — are aligned in exact register
with each other.
- So each chromosome consists of a cable containing 2048 identical strands of DNA.
- These are so large that they can be seen during interphase; even with a low-power light microscope.
What is the function of polyteny?
The probable answer: gene amplification. Having multiple copies of genes permits a high level of gene
expression; that is, abundant transcription and translation to produce the gene products. This would account of polyteny being
associated with large, metabolically active cells (like salivary glands).
Polytene chromosomes are subdivided into some 5,000 dark bands separated by light interbands.
Genes are located in both, but those in the interband regions seem to be more active.
The boundaries between the bands and interbands contain insulators.
Polytene chromosomes also have regions — called "puffs" — that are swollen and appear to have
a looser structure.
The exact pattern of puffs
- differs in different types of cells (e.g., salivary gland vs. gut)
- differs with changing conditions in one type of cell. For example, giving the molting hormone ecdysone to an insect causes a predictable change in the puffing pattern.
This is just what one would predict if the puffs represent regions of intense gene transcription.
 Top: the autoradiograph shows the puff in chromosome IV of the tiny fly Chironomus tentans after giving
ecdysone and radioactive uridine (an RNA precursor). The grains clustered over the puff show that it is a site of intense RNA synthesis. Top: the autoradiograph shows the puff in chromosome IV of the tiny fly Chironomus tentans after giving
ecdysone and radioactive uridine (an RNA precursor). The grains clustered over the puff show that it is a site of intense RNA synthesis.
Bottom: When the antibiotic actinomycin D is given along with ecdysone, puffing is inhibited and little RNA
synthesis occurs. Actinomycin D blocks access to DNA by RNA polymerase. (Images courtesy of Claus Pelling, Max-Planck Institute
of Biology, Tübingen.)
The pattern of puffing within a cell varies over time. For example, each time an insect larva prepares to
molt, a definite, predictable sequence of puffing occurs.
These eight photomicrographs (courtesy of Dr. Michael Ashburner, University of Cambridge) show the changes
in the puffing pattern of equivalent segments of chromosome 3 in Drosophila melanogaster over the course of some 20 hours of normal development.

Note that during this period, when the larvae were preparing to pupate, certain puffs formed, regressed, and
formed again. However, the order in which they did often differed. For example, in the larva, band 62E becomes active before
63E (c, d, and e), but when pupation begins, the reverse is true (g, h).
In general, early puffs reflect the activation of genes encoding transcription factors. These proteins then bind to the promoters of other genes, turning them on and causing a puff to appear at their
loci
Transcription
For a given gene, only one strand of the DNA serves as the template for transcription. An example
is shown below. The bottom (blue) strand in this example is the template strand, which is also called the minus (-)
strand,or the sense strand. It is this strand that serves
as a template for the mRNA synthesis. The enzyme RNA polymerase sythesizes an mRNA in the 5' to 3' direction complementary
to this template strand. The opposite DNA strand (red) is called the coding strand, the nontemplate strand, the plus (+) strand, or the antisense strand.
The easiest way to find the corresponding mRNA sequence (shown in green below) is to read the coding, nontemplate, plus (+), or antisense strand directly in the 5' to 3' direction substituting U for T. 5' T G A C C T T C G A A C G G G A T G G A A A G G 3'
3' A C T G G A A G C T T G C C C T A C C T T T C C 5'5' U G A C C U U C G A A C G G G A U G G A A A G G 3'
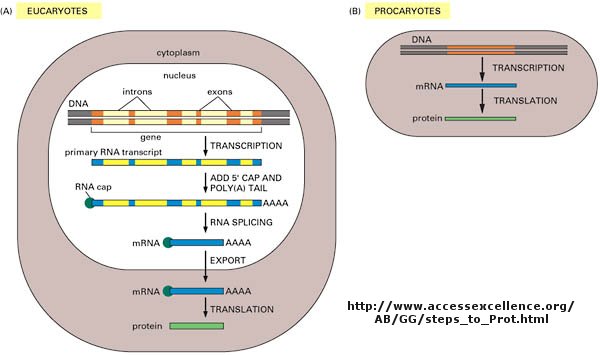
As we've learned more about the structure of DNA, RNA, and proteins, it become clear that transcription
and translation differ in eukaryotes and prokaryotes. Specifically, eukaryotes have intervening sequences of DNA
(introns) within a given gene that separating coding fragments of DNA (exons).
A primary transcript is made from the DNA, and the introns are sliced out and exons joined in a contiguous stretch to form
messenger RNA which leaves the nucleus. Translation occurs in the cytoplasm. Remember, prokaryotes do not have
a nucleus.
Translation
If information in a mRNA sequence is decoded to form a protein. In this process a triplet of nucleotides
(a codon) in the RNA has the information of a single amino acid. Translation occurs on a large RNA-protein complex
called the ribosome. An intermediary transfer RNA (tRNA) molecule becomes covalently linked to a single amino
acid by the enzyme tRNA-acyl synthetase. This "charged" tRNA binds through a complememtary anticodon region to the triplet
codon in the tRNA. The ribosome/tRNA complex ratchets down the mRNA allowing a new "charged" tRNA complex to bind at
an adjacent site. The two adjacent amino acids form a peptide bond in a process driven by ATP cleavage. This process
repeats until a "stop" codon appears in the mRNA sequence. The genetic code shows the relationship between the triplet
mRNA codon and the amino acid which corresponds to it in the growing peptide chain.
Figure: Codon:Anticodon interactions between mRNA and tRNA
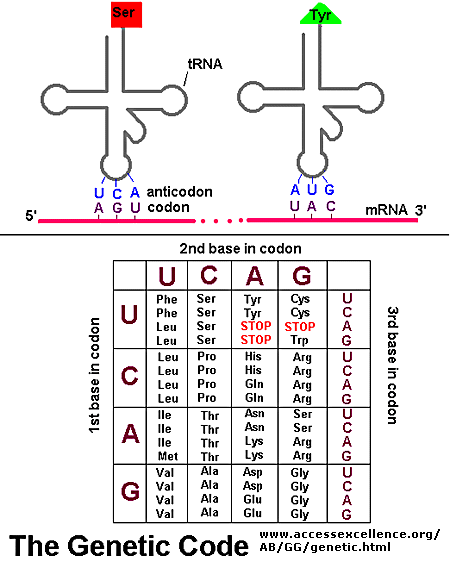
Figure: Central Dogma Differences in Eukaryotes and Prokaryotes
As was mentioned in the Protein Chapter (amino acid section) two
other amino acids occasionally appear in proteins (excluding amino acids altered through post-translational modification.
One is selenocysteine, which is found in Arachea, eubacteria, and animals. The other is just recently found is
pyrrolysine, found on Arachea. These new amino acids derive from modification of Ser-tRNA and probably Lys-tRNA after
the tRNA is charged with the normal amino acid, to produce selenocys-tRNA and pyrrolys-tRNA, respectively. The pyrrolys-tRNA
recognizes the mRNA codon UAG which is usually a stop codon, while selenocys-tRNA recognizes UGA, also a stop condon.
Clearly only a small fraction of stop codons in mRNA sequences would be recognized by this usual tRNA complex. What
determines that recognition is unclear.
YOU CAN SEE A PRESENTATION ON THE CENTRAL DOGMA AT THE SITE http://www.msnusers.com/biochemistryonline
What is a gene?
The definition of a gene can differ depending on whom you ask. The world gene has literally become
a cultural icon of our day. Can our genes explain what it is to be human? The definition of a gene has changed
with time.
Figure: A view of genes and their products: Simplicity to Complexity
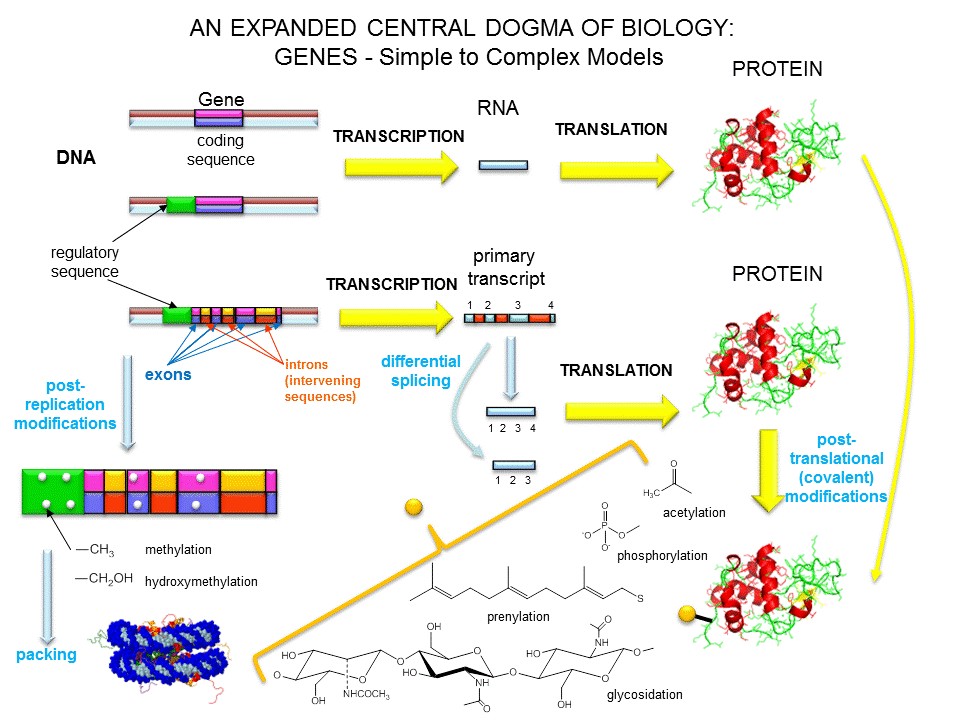
Over the last 100 years, as our understanding of biochemistry has increased, the definition of a gene
has evolved from
- the basis of inheritable traits
- certain regions of chromosomes
- a segment of a chromosome that produces one enzyme
- a segment of a chromosome that produces one protein
- a segment of a chromosome that produces a functional product
The last definition was necessary since some gene products that have function (structural and catalytic)
are RNA molecules. The last definition also includes regulatory regions of the chromosome involved in transcriptional
control. Synder and Gerstein have developed five criteria that can be used in gene identification which is important
as the complete genomes of organisms are analyzed for genes.
- identification of an open reading frame (ORF) - this would include a series of codons bounded by start
and stop condons. This gets progressively harder to do if the gene has a large number of exons imbedded in long introns.
- specific DNA features within genes - these would include a bias towards certain codons found in genes
or splice sites (to remove intron RNA)
- comparing putative gene sequences for homology with known genes from different organisms, but avoiding
sequences that might be conserved regulatory regions.
- identification of RNA transcripts or expressed protein (which does not require DNA sequence analysis
as the top three steps do) -
- inactivating (chemically or through specific mutagenesis) a gene product (RNA or protein).
New Functions for Introns
Recent studies have shown that not all of the DNA interspersed between coding sections of eukaryotic DNA
(introns) are devoid of information content. Some of the intron DNA information ultimately gets transcribed and ultimately
translated into protein structure. Introns with this property can be classifed in two ways:
Group 1 introns: found commonly in yeast, algae, viruses and plant
mitochondria and chloroplasts. After they have been spliced out of transcribed RNA, these RNA sequences ultimately get
translated into proteins with endonuclease activity. These translated proteins differ from typical protein restriction
enzymes which bind and cleave DNA at specific DNA sites of about six base pairs (leading to many cleavages of chromosomal
DNA given the high probability of finding such sites in a large genome). Group 1 protein introns recognize specific
sequences of 15-40 base pairs, which would be found randomly distributed in the genome only once in about 1 billion base pairs.
These Group 1 introns are prime candidates for genetic engineering to produce mutant enzymes which could cut only one specific
site in the human genome, allowing insertion of novel therapeutic DNA sequences into a specific location. This would
alleviate the problem faced in recent DNA therapy regimes in which a therapeutic DNA is inserted into a virus which inserts
randomly into the host DNA, potentially leading to mutagenesis of host DNA. These endonucleases are often called homing endonucleases.
Group 2 Intron also form endonucleases with DNA sequence recognition sites
of 30-35 base pairs. Both the endonuclease and its RNA coding sequences are involved in ultimate insertion of intron DNA (carried on a plasmid) into a specific site in host DNA. The RNA, which is
easier to engineer than proteins, facilitates specific insertion of intron DNA into host DNA. Such insertions can be
used to knockout specific genes with high specificity. (Ingex - with Targetron technology - ge web site). A DNA sequence
can be added by linking a gene of interest to the intron DNA of the plasmid.
Another form of novel "junk" DNA involves the transcription and ultimate translation of a gene whose structure
predicts should be larger than the actual protein which results. Post-translational modification of proteins through
select proteolyis is common. A subset of some mature proteins result from selective removal of an internal piece of the nascent protein
(an intein) from the N terminal (N-extin)and C terminal (C-extein) of the protein and subsequent
religation of the N-extein and C-extein through peptide bond formation. Intein sequence can even be engineered to connect
two different discrete proteins, which is stable at low temperatures, but n which the intein is spliced out at higher temperatures.
The splicing of the N and C-extins occur before the intein is removed. This process occurs during protein folding and
requires no enzymes. The biological role, if any, of inteins is not clear. However, they can be used in the laboratory
in the production and purification of eukaryotic proteins in bacterial whose expression in the absence of an intein produces
a protein that is toxic to the bacterial cell or difficult to purify.
Figure: Mechanism of Intein Splicing
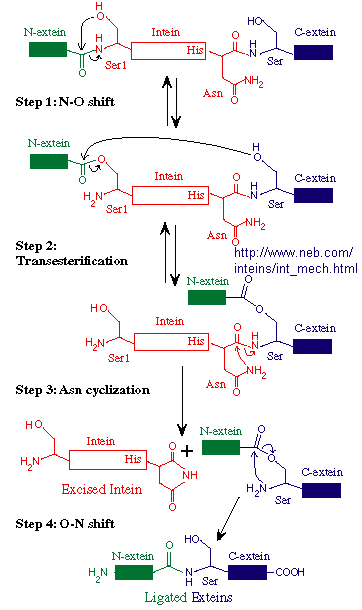
Mutations
Figure: Another View of Mutations
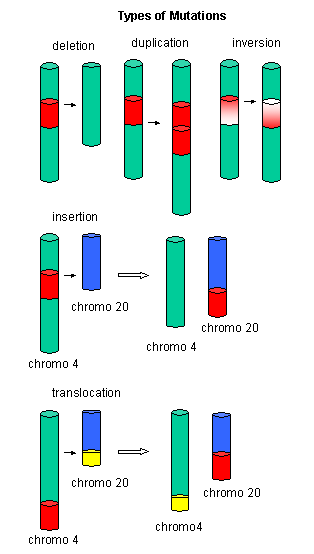
Figure: Point Mutations from: Mismatch pairing and Incorporation of Base Analogs
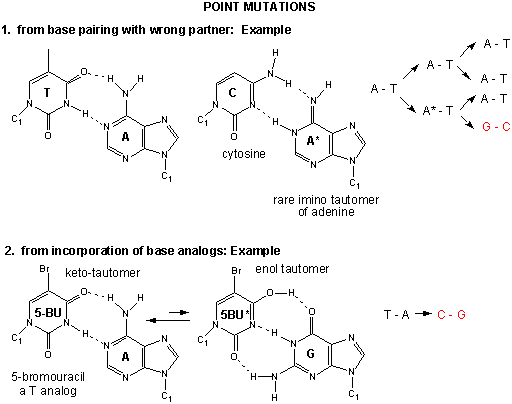
Figure: Point Mutations from: Alkylating Agents

|

